Genomics
All providers authorised to offer sheep genetics testing services working with Sheep Genetics are listed on our website as service providers – you can view our authorised providers here.
What is genotyping?
Genotyping is the process of finding differences in the genetic makeup of individual sheep in their DNA sequences.
DNA is mapped by finding the combination of single nucleotides that make up the DNA or the genome. The genome is the complete map of the individual’s DNA.
These single nucleotide polymorphisms (SNPs) show the differences between your sheep across the genome. At each SNP there are two alleles, one from each parent that indicate differences between sheep.
When you send your blood card or tissue sampling unit to a genotyping company (see bottom of page), they extract the DNA and map these SNPs. This shows what your sheep’s DNA looks like at each of these SNP locations.
The names of these SNPs relate roughly to the number of SNPs each chip has. For example, a 50K chip has roughly 50,000 SNP locations that will be identified.
Genomics to get Australian Sheep Breeding Values
If you subscribe and submit data to Sheep Genetics, your genotyped animals can receive genomically enhanced Australian Sheep Breeding Values (ASBVs). These genotypes estimate how related your animals are to the rest of the animals in the evaluation.
Relationships gives you information for breeding values
ASBVs estimate what genes an animal will on average give to their offspring. Breeding values are estimated based on the traits recorded for an animal and its relatives. Therefore, if your animals are related to genotyped animals in the database, these related animals will contribute information to your animals breeding values. If there is enough information to estimate an accurate breeding value, it will be reported as an ASBV.
These relationships are important for traits that are hard or expensive to measure such as eating quality, traits measured in adults and disease traits.
Collecting more information adds value to the genotypes
Genotyping will help provide information but the best way to get a breeding value is by measuring your own sheep. For more information about how to get involved, contact Sheep Genetics.
Which breeds can genotype to get Genomically Enhanced Breeding Values?
The MERINOSELECT, LAMBPLAN Terminal and LAMBPLAN Maternal are all Single Step evaluations that utilise genomic information. In May 2024 Sheep Genetics introduced an enhancement that enables almost all breeds that have previously submitted genomic tests to have these included in the evaluations to enhance the breeding values provided. This includes many composite and shedding breeds and types. Please note if you are unsure if a breed will have genotypes included in the evaluation please contact Sheep Genetics.
Genomic information is most useful in increasing the accuracy of breeding values where there is an adequate reference population for that group of animals. Reference populations are groups of animals that have both a genotype and phenotype. These are trait specific meaning that a population of animals may have a large reference population for some traits and not others. The size and the relationship to reference population will impact how much benefit a genomic test adds to the predictive accuracy for an animal.
NOTE: the DOHNE evaluation is a non-genomic evaluation meaning genotypes are not included in this evaluation.
MLA genomics database
MLA manage and maintain Australia’s sheep industry genomic database.
This database includes:
- Genotypes from the Cooperative Research Centre for Sheep Industry Innovation (Sheep CRC).
- New genotypes submitted from genotyping companies who signed an agreement with MLA.
MLA own the intellectual property of genotypes from the Sheep CRC and have a license to use new genotypes for parentage verification and analysis.
MLA hold all genotypes in one industry database so:
- genotypes are included in the genetic evaluation for genomic relationships between animals
- existing genotypes in the database can be accessed to assign parentage.
This means whichever company you use, all genotypes can be used together for parentage and genetic evaluation.
What is a flock profile test?
A flock profile test genotypes 20 animals to get an idea of how a flock benchmarks to industry, expressed as ASBVs.
Commercial Merino breeders can use a flock profile test to predicts a flock’s genetics compared to Sheep Genetics data.
How do I get a flock profile test?
Talk to one of the genomics companies that accept genotypes. It’s important to genotype 20 random animals from your flock, from the same year drop.
What do I get from a flock profile?
The flock profile gives average breeding values of a flock for important traits. These numbers are the average estimate for your flock and reflect the genetic merit of your ram team used five to seven years ago.
A flock profile also shows where a flock sits compared to other Merino flocks in the analysis.
Which traits are available?
- yearling fibre diameter
- yearling fibre diameter coefficient of variation
- yearling staple length
- yearling clean fleece weight
- post weaning weight
- yearling weight
- yearling eye muscle depth
- yearling fat depth
- early breech wrinkle
- worm egg count
- yearling fibre curvature
- weaning rate
- condition score
- Fine Wool index
- Wool Production index
- Sustainable Merino index
- Merino Lamb index
What are genetic marker tests?
There are a few genetic conditions or traits controlled by one or a small number of genes. These genes are found using single nucleotide polymorphisms (SNPs).
A marker report include tests for the following:
- yellow fat
- hairy lamb
- callipyge
- myostatin
- spider lamb
- TMEM
- micropthalmia
- scrapie
- texel chondro
- GDF9
- inverdale.
Breeders are advised to seek advice when using genetic conditions and recessive genes in a breeding program. Not all genetics conditions and recessive genes have been validated as functional for all breeds.
How does parentage assignment work?
Parentage assignment uses single nucleotide polymorphism (SNP) information to allocate potential parents.
Potential parents are checked to find who shares at least one allele at each loci. For example, potential parents are ruled out in the purple box because they do not share enough alleles as the animal tested.
Parentage assignment example
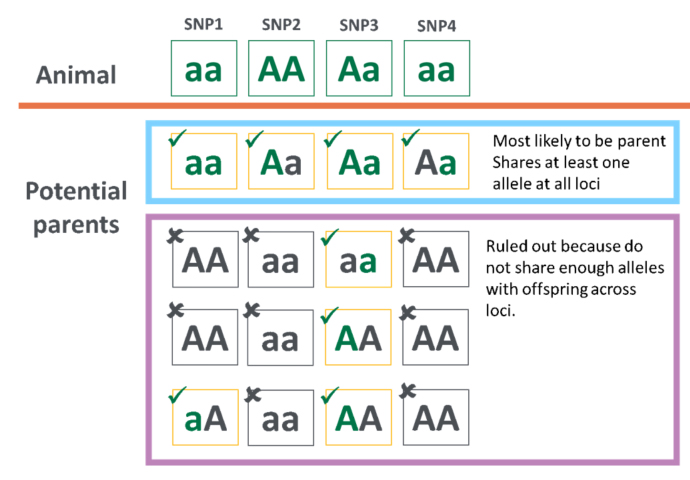
Accurate sire and dam lists make parentage easier
To assign parents, all potential sires and all dams need to be genotyped. This assignment to parents rules out potential sires and dams that do not match the progeny. For this reason, it is important to submit accurate and complete sire and dam lists for the best results.
The genomic coverage matters
The parentage assignment can only use the lowest single nucleotide polymorphism (SNP) coverage available in the potential candidates.
If all offspring are genotyped with 1,200 SNPs but some potential parents are genotyped with old 170 SNP panels, then only 170 SNPs are used for the parentage test.
Genotyping providers
The table below displays the genotyping providers and the products they offer.
| Parentage only | Parentage and genomic test | Flock profile | Marker tests | |
| GenomNZ | X | X | X | X |
| Intertek | X | |||
| NEOGEN | X | X | X | X |
| Weatherbys | X | X | X | |
| XYTOVET | X | X | X | X |
Provider contact details
GenomNZ
Rayna Anderson
P: +64 800 362 522
E: genomnz@agresearch.co.nz
Intertek
Tania Pfeiffer
P: 0459 855 862
E: tania.pfeiffer@intertek.com
NEOGEN
Melanie Dowling
P: 07 37362134
E: naa-sheep@neogen.com
Weatherbys Scientific Australia
Tamarah Luxton
P: 0474 633 799
E: tluxton@weatherbys.com.au
XYTOVET
P: 08 6383 8110
E: samples@xytovet.com.au